Output
Running a BioLib application generates the following outputs:
- Main output is shown to the user in the application window once an application has completed running.
- Runtime output is shown to user in the application window during execution of the application.
- Output files that the application produces can be downloaded by the user.
- Logs that contain information about the runtime process and potential errors.
Main output
In the main application window, upon completion, the following output will be shown:
- If the application specifies
main_output_filein the.biolib/config.yml(typically Markdown or HTML file) this file will be rendered. - If the application does not specify
main_output_filethe stdout will be rendered.
The main output file is specified in the .biolib/config.yml, as main_output_file: your_file.md . You can read more
about the syntax of the config.yml file here.
If the main_output_file is a markdown file, this will be rendered in the application window and can be used to generate
reports with descriptions, tables, and figures.
HTML - Introduction
You can set main_output_file to point to an HTML file in the output files of your application. This allows you to
create interactive user experiences such as dashboards and plots for analyzing the results of the application.
The HTML file will be rendered inside an iframe and have access to a JavaScript API for retrieving and storing data.
For security reasons, the iframe will not have internet access and thus cannot load remote dependencies such as JavaScript libraries or stylesheets. Therefore, you must bundle / embed any dependencies that you require inside the HTML file.
HTML - JavaScript API
When your HTML file is loaded into the iframe, a special API class called biolib is loaded onto the browser's window
object. The window object is globally accessible and thus the API class can be called from any part of your code. For
instance, you could call window.biolib.listOutputFilePaths() to get a list of all the output files.
The full definition of the API class is shown below:
const biolib: {
// Methods
getInputFileData: (path: string) => Promise<Uint8Array>;
getOutputFileData: (path: string) => Promise<Uint8Array>;
getFileDataFromDataRecord: (options: {uri: string, path: string}) => Promise<Uint8Array>;
listOutputFilePaths: () => Promise<string[]>;
queryIndex: (payload: object) => Promise<object>;
runJob: (payload: {appUri: string, arguments: string[], files: {[key: string]: {data: Uint8Array}}, blocking?: boolean}, jobProgressHandler?: (progress: {jobId: string, logMessage?: string, systemLogMessage?: string}) => void) => Promise<Result | PendingResult>;
// Properties
filePath: string;
job: null | {id: string, authToken: string, appUri: string};
}Methods
getInputFileData(path) retrieves the content of an input file that was provided to the application.
getOutputFileData(path) retrieves the content of an output file that was generated by the application.
getFileDataFromDataRecord(options) retrieves file data from a DataRecord. The options object must contain uri (the DataRecord URI) and path (the file path within the DataRecord).
listOutputFilePaths() returns a list of all output file paths available.
queryIndex(payload) queries the BioLib index with the provided payload.
runJob(payload, jobProgressHandler) calls another BioLib application. When blocking is false, it returns immediately after job creation with {jobId: string, authToken: string} instead of waiting for the job to complete. This allows you to start a job and redirect to the results page. The optional jobProgressHandler callback receives progress updates during job execution.
Properties
filePath contains the path of the current file being rendered (if applicable).
job contains information about the current job context, including id, authToken, and appUri. This is null if no job context is available.
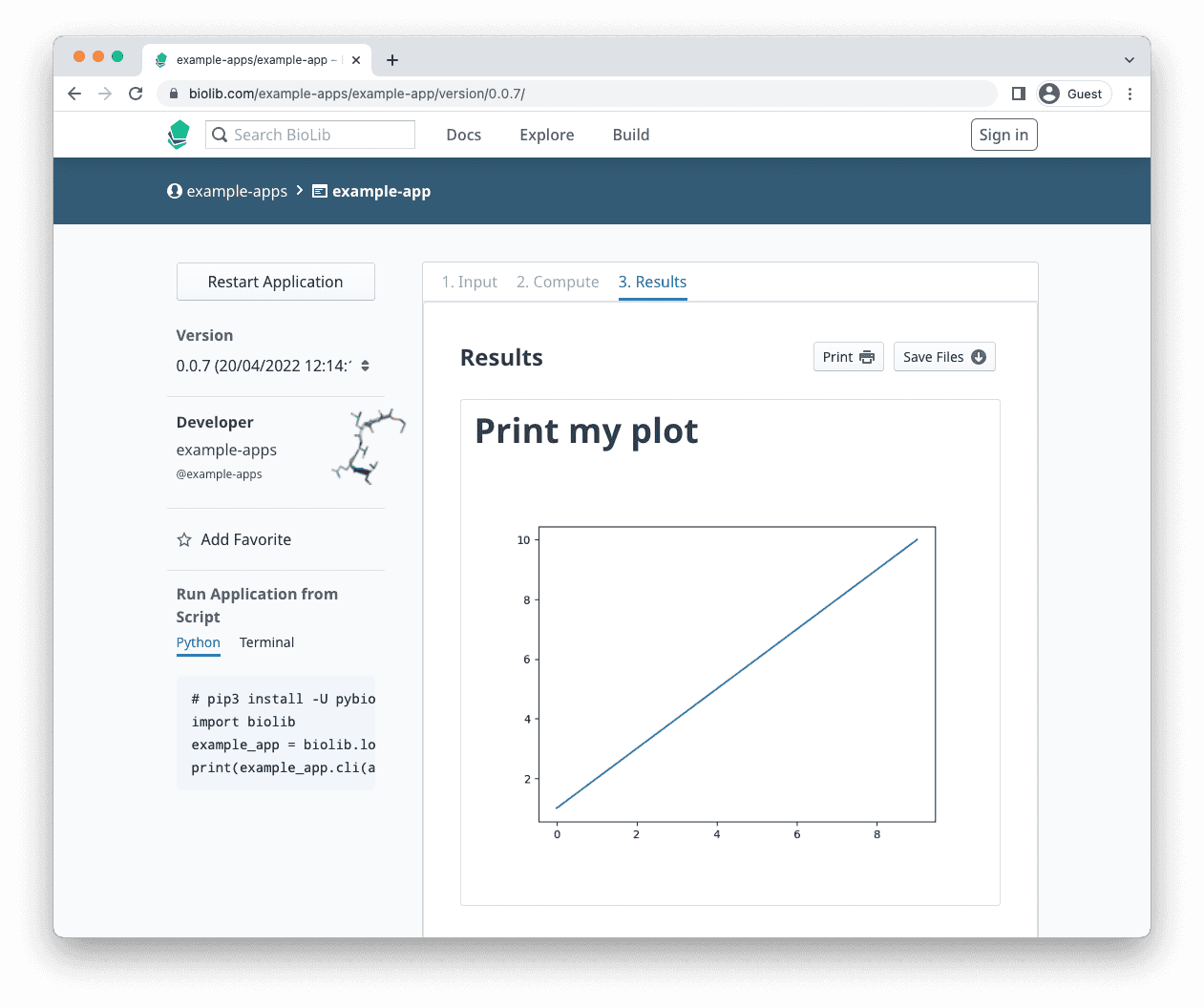
Markdown Example: Show figures
The figure my_plot.png can be shown in the main output file by using the Markdown syntax as shown below.
# define imports
import matplotlib.pyplot as plt
# generate plot
plt.plot([1, 2, 3, 4, 5, 6, 7, 8, 9, 10])
plt.savefig('my_plot.png')
# Generate a main output file
with open('output.md', mode='w') as output:
# add a title
output.write(f'# Print my plot\n\n')
# add the figure
output.write('')
Runtime Output
During execution of an application, end-users will see the runtime output and the application log.
The runtime output will show any messages that the application prints or writes to output during the execution of the applications. For example, the python application below takes an integer input and prints the range from 0 to the input integer, which will be shown as the runtime output.
# import libraries
from time import sleep
import argparse
# read inputs provided by user
parser = argparse.ArgumentParser()
parser.add_argument('--digit', type=int)
args = parser.parse_args()
# run application functions
print('Step 1 | Print digits')
for i in range(args.digit):
sleep(.25)
print(i)
print('I have finished printing all the digits!')
# print output.md
print('Step 2 | Write output')
with open('output.md', mode='w') as output:
output.write(f'# This is an example\n\n')
output.write(f'This app wrote {args.digit} digits')The runtime output is rendered as plain text, and when an application has finished computing the stdout will be viewable in the runtime output log.
Output files
Once the computation has completed, all files that have been written to disk are saved to a virtual filesystem. All
files written to disk, as well as stdout and stderr, can be downloaded by the end-user by clicking the download
button in the GUI or using the function results.save_files('your_directory/') from Python (
see Running applications in python).
Example: Writing to disk
Any new file created or edited during computation will be available for the user of the application to download once the
computation has completed. In the Python example below, the application creates a file called my_name.txt which can
be downloaded by the user.
import argparse
# read inputs provided by user
parser = argparse.ArgumentParser()
parser.add_argument('--name')
args = parser.parse_args()
# write output to file
with open('my_name.txt', mode='w') as outfile:
outfile.write(f'My name is {args.name}')While the format and rendering of outputs to the main output are limited to text, Markdown, and HTML, outputs written to disk can use any file format, like FASTA, FASTQ and other formats commonly used in bioinformatics.
Logs
A BioLib application produces two types of logs.
- The application log contains information about how the application is being run.
- Standard Output and Standard Error contain the stderr and stdout from the application.
These files are written as hidden files
.stderrand.stdoutin the output files.
Name the result at runtime
The name of the result the user will see from running your app can be set a runtime with below:
from biolib.sdk import Runtime
Runtime.set_main_result_prefix('my-example-result')Still have a question?
If you have any questions that you can't find an answer to above, please reach out to the BioLib community.